An extensive set of tools supports fixing imaging artifacts in single-frame and time-lapse images, that would otherwise interfere with image analysis and quantification. These functions may save for use troubled recordings, e.g. a sample that moved during time-lapse recording, or bestow automated image analysis with robustness. Using motorized stage and multi-position time-lapse recordings necessitates using image registration algorithms even with the most precise linearly encoded stages. All image registration routines use high speed, multi-core CPU optimized DFT routines, and image transformations with the choice of interpolation. All of these functions retain intensity values so images remain fully quantifiable.
Image Stabilizer (Image registration in time lapses)
The algorithm automatically detects and corrects frame-to-frame dislocations of the content of the images in time lapses, resulting in a steady time series. The Image Stabilizer corrects for the frame-to-frame jitter caused by the inaccuracy of stage motors in Multi-Dimensional experiments or for slow drift of the sample which develops in a span of many frames. The tiled version of this function corrects for incorrect stitching in time lapses.
To perform image registration (stabilization)
Load a single or multi channel image recording
Select the
![]() Align
Series (Image Stabilizer) function in the Editing
menu
Align
Series (Image Stabilizer) function in the Editing
menu
Select the best signal to noise ratio channel as Image (A) by checkmarking in the left list in the center of the parameter bar
Select all images as Image (B) by checkmarking in the right list in the center of the parameter bar
The image registration is ready run now by
pressing the
![]() button
button
To tune the Image Stabilizer, repeat points 1-4 and adjust the following parameters before processing images
Set working size as 2^working size is greater than magnification factor times the width (or height) of the image in pixels
Set compare to the first frame Yes, if the imaged objects do not change much during the recording
If not comparing to first frame, use difference period 1 for correcting jitter, or larger values, e.g. 3-5 for slow movement of the image
Align Series corrects for an xy-offset of the image, but not for rotation. If the image rotates, use the Manual alignment (Main Menu/Edit/Manual Image Stabilizer).
Select Main Menu/Edit/Manual Image Stabilizer
Right-click the image to show keyboard shortcuts for transformation of the image, and register the red image with the green (first frame) for critical key frames.
Each transformed frame sets up a key frame (marked by an L in the top left corner, and the transformation of each frame (red) is interpolated in between keyframes.
Press Enter to transform this image series or all linked channels.
Related functions:
Example for Image Stabilizer
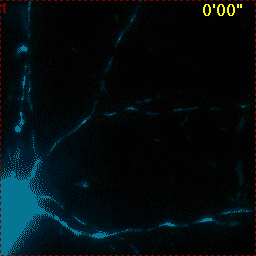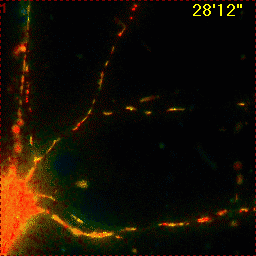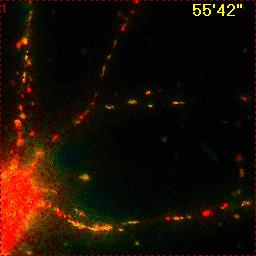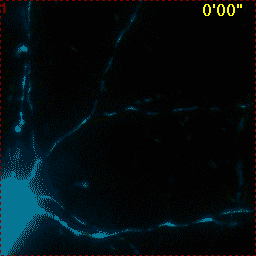 |
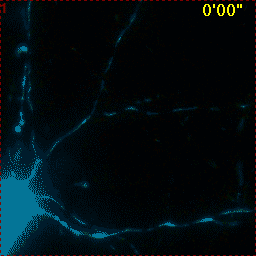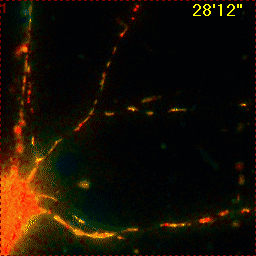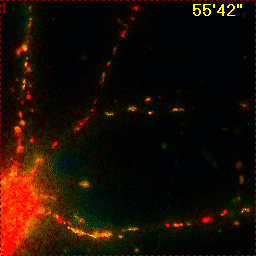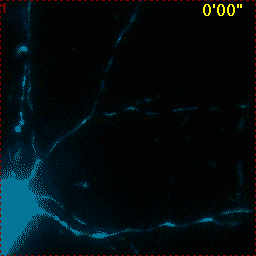 |
Hippocampal neuron expressing mitochondrially targeted yellow cameleon 4mtD3cpv. FRET ratio time lapse shows the effect of glutamate stimulation. The multi-stage position recording caused substantial shaking of the image, that is corrected by image registration. |
| Original |
Using
|
Z-drift stabilizer (beta)
Image Analyst MKII is a 2D time lapse analysis software, but supports analysis of XYZT stack by calculating z-projection during reading the data set. Optionally a sub-z-stack can be projected, and the center plane of this sub stack can follow frame-to-frame drift in z. To this end in the Multi-Dimensional Open dialog/Setting tab click "Use z-drift stabilizer" , and configure Z-stabilizer on the corresponding new tab.
Channel Alignment (image registration between channels)
The algorithm automatically detects and corrects lack of registering multiple fluorescence channels at subpixel level. Typical use is correction for alignment errors originating from using different dichroic mirrors and filters for recording different channels. Alignment is also possible between different, not linked image sets in order to register different recordings, e.g. live cell imaging with post-hoc immunostaining. Tiled version corrects for incorrect stitching if different channels were recorded by successive tilings.
To perform channel alignment
Load a multi channel image recording (or two recordings if aligning between recordings)
Select the
![]() Align Channels
function in the Editing menu
Align Channels
function in the Editing menu
Select the channel (recording) to be transformed as Image (A) by checkmarking in the left list in the center of the parameter bar
Select the channel to compare to as Image (B) by checkmarking in the right list in the center of the parameter bar
If the alignment is between two recordings, optionally set "Transform all linked images" to Yes to align all channels of the first recording (A) to to the second recording (B).
The image registration is ready run now by
pressing the
![]() button
button
To tune the Channel alignment, repeat points 1-4 and adjust the following parameters before processing images
Set working size as 2^working size is greater than magnification factor times the width (or height) of the image in pixels
Related functions:
Spectral Unmixing
Algorithms are available for calculation of crossbleed coefficients and for performing linear unmixing. Adjustments for differing exposure times used for coefficient measurement and recordings to be unmixed are possible.
Related functions:
Correction for sudden changes in illumination intensity
Lens distortion, spherical aberration
Wavelength-dependent lens distortion may preclude correct colocalization of objects. These functions allow correction by un-distorting (straightening) images:
Related functions:
Removing debris, dead cells
Using image segmentation and masking unwanted details, fluorescent debris, washed-off cells, dead cells, recodings can be pre-processed before analysis. Typically a single binary image is calculated by maximum intensity projection of the time lapse, segmentation (e.g. looking for bright round dead cells), thresholding (creating a binary mask), and masking the whole time lapse with the binary mask.
Related functions:
Updated: 09/23/2015