Fluorescence measurement of mitochondrial swelling in cells
The "ratiometric
optimized spatial filtering technique" or
"thinness ratio" (TR) technique
measures subtle, subpixel changes of diameters of
fluorescent objects in microscopic images. The
relative contribution of fluorescence intensities of
thinner versus thicker punctate or filamentous
structures is measured by a set of two band pass
spatial filters. The protocol given below provides a
quantitative, single cell, in situ
measurement of mitochondrial swelling. The TR
decreases proportionally with the increase of the
diameter of mitochondria during mitochondrial
swelling and is specific to mitochondrial swelling
as compared to mitochondrial fission. The technique
is sensitive to diameter changes at a 10 nm scale.
The exact description of the technique can be found
in
Gerencser et al. 2008.
Mitochondrial swelling measurement with the TR technique consists
of a calibration recording, looking the effect of valinomycin, and
arbitrary number of experiments to be calibrated. The calibration
recording is performed per microscopic setting and type of
biological specimen (e.g. different fluorphores). Experiments
belonging to the same data set have to be recorded under identical
microscope settings and processed with the same filter functions to
ensure unbiased data.
This protocol assumes that the user is familiar with the
following sections of the online manual:
Important imaging and specimen requirements:
- TR is calculated from high resolution wide-field or confocal
microscopic grayscale images visualizing fluorescently marked
mitochondria.
- The specimen must stay completely in focus. Even slight
focus drift can cause decaying TR. If the specimen is thick or
the focus drifts, use z-stacking. Thus the thickness and
drift determines the number of z-steps, simply to keep the
observed mitochondria within the imaged volume. However, TR can
be measured in single plane images, if the focus is stabile
enough.
- The fluorophore must not redistribute. Therefore membrane
potential dyes (TMRM, TMRE, Rh123), Mitotrackers, NOA are
unsuitable for TR analysis. Use targeted fluorescent proteins.
- The detector must not be saturated. Edges of saturated areas
contain abnormally high frequency components.
- For the fastest analysis use quadrangular, power of two
sized images, ideally 512x512.
|
|
Important parameters
|
Additional parameters
|
|
Fluorophore:
|
large molecular weight,
organelle specific, bright
|
l is less important,
can be a probe
|
|
Focus:
|
z-stacking and mean
intensity projection
step size =
axial resolution ~0.8
mm
5-7 steps depending specimen thickness and focus
drift
|
Single image plane can be also used if the
constant focus can be maintained e.g.
-TIRF nosepiece e.g. Olympus
-active autofocus: e.g. Nikon Perfect Focus
System, Zeiss LSM Multi Time Lapse Module
autofocus
|
|
Optics:
|
NA>0.8, but higher is
better; if confocal, pinhole>1.5
resel (or Airy unit)
|
confocal or wide-field, latter is
preferred
|
|
Detector:
|
0.1-0.14 mm/pixel
resolution
~1000 photo e-/pixel
(summed for the z-stack)
DO NOT SATURATE
|
Image is qudrangular, size is power of 2,
ideally 512x512. If using CCD cameras use
sub-frame readout |
|
| Modified from
Gerencser et al. 2008.. |
Recording of the calibration image set
- Use microscope settings as above given. The calibration and
to-be-calibrated recordings must use identical settings.
- Record baseline for at least 3 time frames
- Add valinomycin (200 nM) to the sample and when it's
swelling-inducing effect is visible record at least further 3
time frames.
Recording arbitrary image sets (experiments)
- Use the same microscope settings as above. If using
different settings (e.g. different magnifications), the
calibration has to be performed.
Performing optimization of filter functions on the
calibration image set
- Load the calibration image sequence (download tutorial data
set:
 ).
).
- To z-project XYZT stack the input format must be *.nd,
*.nd2 or *.lsm. In the
Multi-Dimensional Open dialog Settings tab
set Project Z to Mean
Intensity. The checkboxes in the Settings tab
remain unchecked, unless partial reading of the image stacks
is to be performed. For loading non-Multi Time Lapse *.lsm
files the z-projection is set in the Preferences
Data/Loading/ROIs tab.
- For other formats z-project z-stacks before loading into
Image Analyst. Use mean intensity projection. If it's not
available, use maximum intensity projection. Export as tif-files.
Load a set of tif-files into the Image Analyst by multiple
selection.
- Truncate image sequence to contain 3 (or more) baseline
followed by 3 (or more) valinomycin treatment frame using the
Truncate Image Series
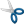 .
Larger number of frames will take longer time to optimize. In
the tutorial data set this is the first and last 3 frames.
.
Larger number of frames will take longer time to optimize. In
the tutorial data set this is the first and last 3 frames.
- Draw a ROI encircling mitochondria which stay in focus
during the experiment, and the valinomycin-triggered swelling is
well visible.
- Optionally create a mask image. Mask image will improve
signal to noise ratio by supressing background. Never use
masking on the original image.
-
 Copy the image, linked.
Copy the image, linked.
- By using the
 Segmentation/Threshold
function
binarize the copy.
Segmentation/Threshold
function
binarize the copy.
- Threshold value calculation method:
Otsu by frames
- Way: Above
- Value: 1 (decrease this value to have
more mitochondria or increase to have less interference by
background noise)
- Determine boundaries at: 0
(this number is indifferent here)
- Threshold from local max/min:
None
-
 Process
e.g. by using the
context menu of the Image Window.
Process
e.g. by using the
context menu of the Image Window.
- Open the Setup DFT Filter
dialog from the Tools main menu
- In the Calibration tab enter
the Spatial resolution (pixel size) of the image. This can
be determined either by pressing Ctrl-I invoking image
information in an Image Window, if spatial resolution was
saved into the image by the image acquisition software.
Alternatively determine it in your image acquisition
software. For CCD cameras spatial resolution can be
calculated as binning * physical pixel size / lens
magnification / any additional zoom, optovar. This value is
0.16 mm/pixel in the tutorial
data set.
- Note the maximal w,
which is calculated from the spatial resolution by the Image
Analyst.
- In the Optimization tab select the Image to be
Optimized. If using mask image, select the mask image as
well, otherwise disregard. If the image names do not appear,
switch between the Calibration and Optimization
tabs back and forth.
- At the Select parameters of
select Differential Evolution Optimizer.
For the tutorial, proceed to next step.
- The optimizer searches for the filter functions. The
default settings result robust optimization of filter functions in mitochondrial swelling assays in cortical neurons and astrocytes imaged at
~0.1 mm/pixel resolution. In the case of misconvergence increase the value for "Population
size" or adjust the "Weight" to be a little lower or higher than 0.8. If you increase "Population
size" and simultaneously lower "Weight" a little, convergence is more likely to occur but generally takes longer. High values of "Crossover"
like 1 give faster convergence if convergence occurs.
However, you may have to go down as much as CR=0 to make
DE robust enough.
- At the Select parameters of
select Thinness Ratio Filter Pair.
For the tutorial set only the maximum cutoff values (point
2-3 below) and the Use Mask (point 10), and proceed to next
step.
- The units of
spatial frequency is given in cycles/mm
as default, but can be changed at the Spatial freq. (ω) unit.
- The low band pass (LBP) filter will be searched
between LBP minimum cut on
ω and LBP maximum
cut off
ω values typically
between 0-1 cycles/mm (see
Gerencser et al. 2008 Fig.2.).
The maximal value must be below the value of maximal
w determined at point
5.2 above.
- The high band pass (HBP) filter will be searched
between HBP minimum cut on
ω and HBP maximum
cut off
ω values typically
between 1-3 cycles/mm (see
Gerencser et al. 2008 Fig.2.).
The maximal value must be below the value of maximal
w determined at point
5.2 above.
- The order of the filter functions (same for all cut
ons and cut offs) is searched between Order minimum
and Order maximum. Higher order results steeper
functions.
- Filter normalization:
typically corrected integral (see
Gerencser et al. 2008 Eq.5,). is used to result
intensities at similar order of magnitude after
filtering, however as Image Analyst MKII is fully 32bit
floating point based, it has little importance.
- Test ROI :
1 (if there was only on ROI on the
image) Set the number of the ROI to be analysed.
- Number of baseline frames:
3 (The image sequence consists of the
given number of baseline frames followed by the given
number of treated frames. The set of baseline and
treated frames can be repeated any time in the same
order with the same number of frames to increase the
power of the statistics. In the tutorial data set this
value is 3. )
- Number of treatment frames:
3 (This is the number of frames
corresponding to the valinomycin treatment. In the
tutorial data set this value is 3.)
- Direction of change:
Decreasing (The optimizer can look for
an increasing or a decreasing signal, this is decreasing
if valinomycin treatment follows the baseline, as
mitochondria are expected to swell and the TR value to
decrease.)
- Use Mask: Yes
if having a separate mask image. No
if not. TR will be calculated only at those areas where
the mask image has values of '1'.
- Preserve edges: No
(Performs mirrored tiling to prevent edge artifacts.
Slows down processing by 4x. Set it to Yes
if interested in details close to the edge of the
image.)
- Enlarge paper: No
(Enlarges image by mirrored tiling to quadrangular and
prevents edge artifacts and distortions from
non-quadrangular images.) Set this to Yes
if working on non-quadrangular image.
- Enlarge to 2^: 9
Size of image during filtering as power of 2. Used only
if Enlarge paper is set to Yes.
- Leave only phase:
No
- Absolute:
Yes
- Protect
MASK:
No (Fills up masked areas and
undetermined pixel values with zeros before processing.
Required for masked images. In this case the image is
not expected to contain masked areas).
- Press Optimize, and wait for convergence which
can take several (5-10) minutes.
- The results for each iteration step are given in the
Results box. The results are given as LP or HP cut
on-cut off, order; using pixels as
spatial
frequency units. This avoids the requirement of the
image resolution data for further processing. Write
down the last pair of LP and HP values.
 |
| The results of the optimization are given in the
Results box on the right. |
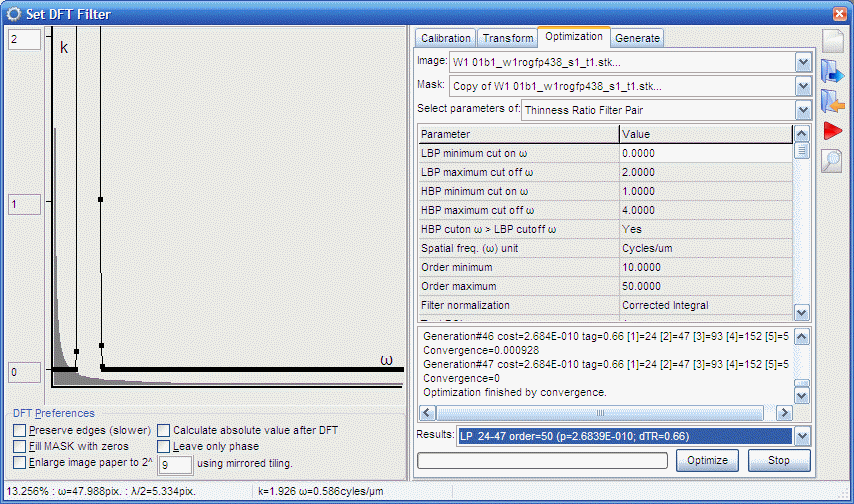
Creating and using a pipeline to perform TR calculation in an
arbitrary image set
 |
| Mitochondrial Swelling by Thinness Ratio.ips |
- Open a new Pipeline
window (main menu Pipeline) and open My Documents/Image
Analyst/macros/Mitochondrial Swelling by Thinness Ratio.ips
- Click the left 2D DFT Butterworth BP filter in the
block diagram, this will be the low band pass (LBP) filter.
- Cut On
w: enter here the first
number after the LP in the results box above
- Cut On order and
Cut Off order: enter both places the
order after the LP in the results box above
- Cut Off
w:
enter here the second number after the LP in the results
box above pixels
- unit of
w:
pixels
- The rest of the parameters are set identically as it
was done in the Optimization/Thinness Ratio Filter Pair
above, points 5.5.10-5.5.16. Using Absolute
value calculation (Yes) is critical.
- Click the right 2D DFT Butterworth BP filter in the
block diagram, this will be the high band pass (HBP) filter.
- Cut On
w:
enter here the first number after the HP in the results
box above
- Cut On order and
Cut Off order: enter both places the
order after the HP in the results box above
- Cut Off
w:
enter here the second number after the HP in the results
box above pixels
- unit of
w:
pixels
- The rest of the parameters are set identically as it
was done in the Optimization/Thinness Ratio Filter Pair
above, points 5.5.10-5.5.16. Using
Absolute value calculation (Yes)
is critical.
- The Threshold in the middle has to set similarly as it
was used for masking during optimization
- The 2D Nonlinear filters (maximum
filters) are necessary for TR image creation. However for
plotting only (
 Ratio
Plot) TR data, these filters should be discarded from the
pipeline.
Ratio
Plot) TR data, these filters should be discarded from the
pipeline.
- To erase 2D Nonlinear filters, unlock editing
 ,
,
- Right-click 2D Nonlinear filter and select Delete.
- Drop The Apply mask by division
first on the 2D DFT Butterworth filter,
then on the Threshold, to maintain correct order of
inputs.
- Save the pipeline under a new name.
- Load an arbitrary image sequence (e.g. the complete image
sequence of the tutorial - 9 frames). Use the same kind of
projection as for the calibration.
- To z-project XYZT stack the input format must be *.nd,
*.nd2 or *.lsm. In the
Multi-Dimensional Open dialog Settings tab
set Project Z to Mean
Intensity. The checkboxes in the Settings tab
remain unchecked, unless partial reading of the image stacks
is to be performed. For loading non-Multi Time Lapse *.lsm
files the z-projection is set in the Preferences
Data/Loading/ROIs tab.
- For other formats z-project z-stacks before loading into
Image Analyst. Use mean intensity projection. If it's not
available, use maximum intensity projection. Export as tif-files.
Load a set of tif-files into the Image Analyst by multiple
selection.
- Process images by running the pipeline using the
 button.
button.
 |
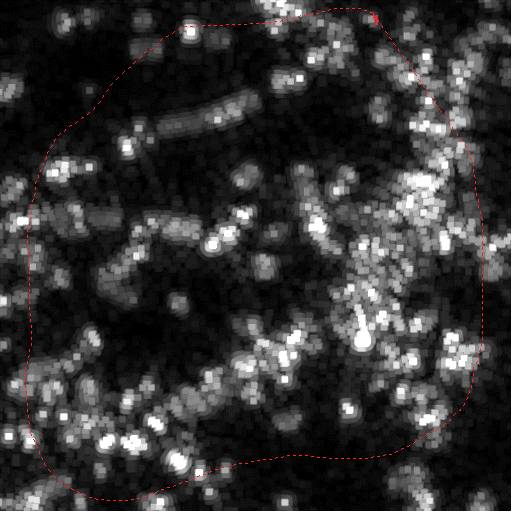 |
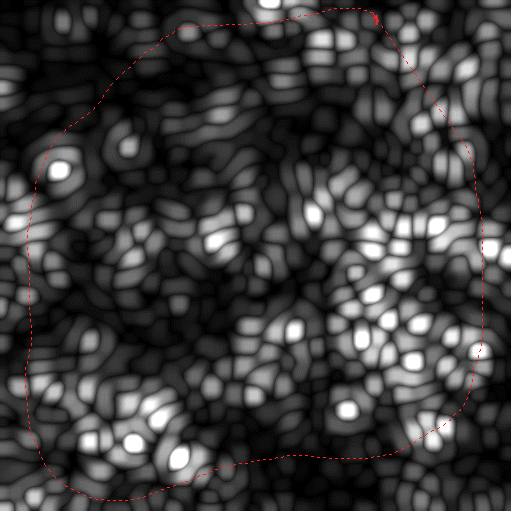 |
|
Raw, mean intensity projected image with test ROI |
HBP and maximum filtered image before masking |
LBP and maximum
filtered image before masking |
 |
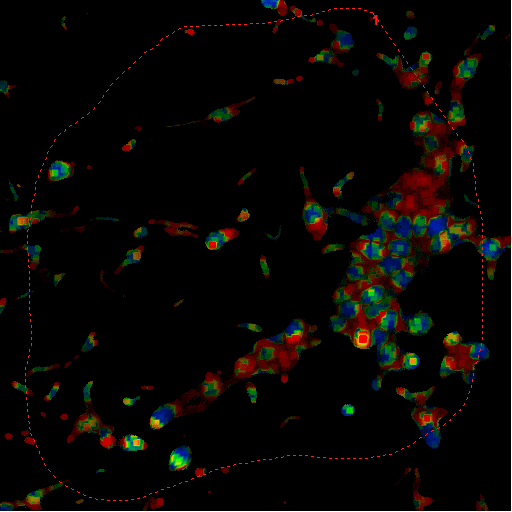 |
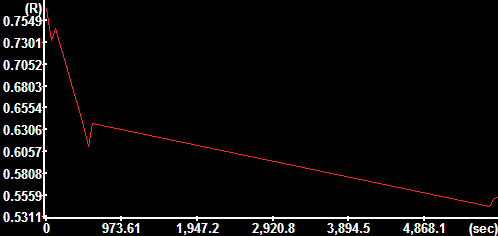 |
|
mask image |
TR |
TR data |
Notes:
- TR is insensitive to fluorescence background as long as the
cut ons are larger than zero. Therefore no background
subtraction is required.
Protocol by Akos A. Gerencser 02/03/2010 V1.0
References
1.
Gerencser
A. A., Doczi, J., Töröcsik, B., Bossy-Wetzel, E., and Adam-Vizi,
V. (2008). Mitochondrial Swelling Measurement in Situ by Optimized
Spatial Filtering: Astrocyte-Neuron Differences. Biophys
J.Sep;95(5):2583-98.







![]()