Measurement of velocities and fluorescence intensities over single
mitochondria
This protocol describes how to segment fluorescence
time lapse image sequences to determine velocities and
other parameters of individual mitochondria, like
intensities of other fluorescence channels, size and
length. The technology described here is an improved
version of our earlier described
method. The protocol consists of the
preparation of images for segmentation (local background
removal, smoothing), segmentation and reading out values
corresponding to segments. Measurement of anterograde
and retrograde or directional vs. wiggling motion is
given at the bottom.
This protocol assumes that the user is familiar with the
following sections of the online manual and protocols:
Protocol for segmentation
- If using Multi-Dimensional Open dialog:
- Load the original (raw) fluorescence time lapse image sequence
with minimum intensity projection (Settings tab).
Minimum intensity projection is used to show only those
mitochondria that move slowly, thus the Optical Flow and
intensity readouts will be correct.
- Optionally, subsequently, load other channels, if
present, e.g. the TMRM channel in the example below.
- Perform Optical Flow load as described
in the
"Organelle motion assay with Optical Flow".
- If using simple time lapse recordings, load the original
(raw) fluorescence time lapse image sequence.
-
 Create
a copy of the image.
Create
a copy of the image.
- Perform Optical Flow on the copy as described
in the
"Organelle motion assay with Optical Flow".
- Rename the resultant image to 'Optical Flow' (Main
menu/Edit/Rename).
- Project the original image with the Filters/
 Temporal
block filter / Filter type=Min / Window width = the
value of SG kernel for dt width or 2 if dt kernel=[1,-1]
for the Optical Flow calculation. The value of
Block mode is identical to the Block mode of
the Optical Flow. Minimum intensity projection is used to
show only those mitochondria that move slowly, thus the
Optical Flow and intensity readouts will be correct.
Temporal
block filter / Filter type=Min / Window width = the
value of SG kernel for dt width or 2 if dt kernel=[1,-1]
for the Optical Flow calculation. The value of
Block mode is identical to the Block mode of
the Optical Flow. Minimum intensity projection is used to
show only those mitochondria that move slowly, thus the
Optical Flow and intensity readouts will be correct.
- Link images with
 .
.
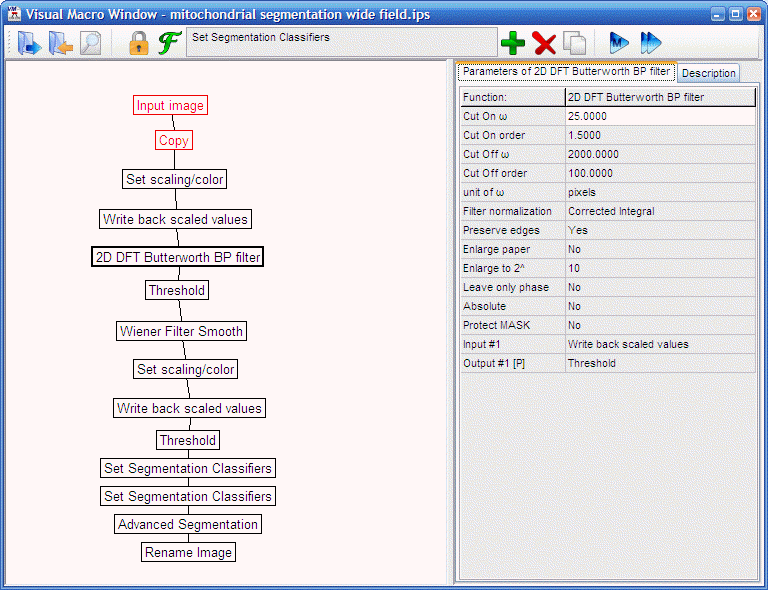
- Load the "mitochondrial segmentation wide field.ips" ,
select the minimum intensity projection image and use the
 to run it (the steps of the pipeline are detailed below, in routine
use proceed to the next step):
to run it (the steps of the pipeline are detailed below, in routine
use proceed to the next step):
-
 Set
scaling/color and
Set
scaling/color and
 Write
back scaled values: values between 0-99.99
Write
back scaled values: values between 0-99.99
-
 2D
DFT Butterworth BP filter: 25/1.5/2000/100/pixels/.../absolute=No :
this is a highpass filter with a cuton at 25 pixels spatial
frequency to show mitochondria only. Note that if images with
different size or spatial resolution are to be processed, a different cuton frequency will be
required. See more in
"Spatial filtering in Fourier domain"
and in "Background removal by highpass filtering". The spatial frequency
value of 25 pixels is 0.175 cycles/mm
at 512x512 images at 0.28 mm/pixel
resolution in the example shown below. The 0.175 cycles/mm
is a constant value for mitochondrial filtering. So if using
different image size or resolution, set the spatial resolution
in the Tools/
2D
DFT Butterworth BP filter: 25/1.5/2000/100/pixels/.../absolute=No :
this is a highpass filter with a cuton at 25 pixels spatial
frequency to show mitochondria only. Note that if images with
different size or spatial resolution are to be processed, a different cuton frequency will be
required. See more in
"Spatial filtering in Fourier domain"
and in "Background removal by highpass filtering". The spatial frequency
value of 25 pixels is 0.175 cycles/mm
at 512x512 images at 0.28 mm/pixel
resolution in the example shown below. The 0.175 cycles/mm
is a constant value for mitochondrial filtering. So if using
different image size or resolution, set the spatial resolution
in the Tools/ Setup DFT Filter
dialog Calibration tab,
and either calculate
the new cut on spatial frequency in pixels as 0.175/w
resolution (w
resolution is displayed below the spatial resolution) or
set the 2D DFT Butterworth BP filter the
unit of w parameter to
cycles/mm.
Setup DFT Filter
dialog Calibration tab,
and either calculate
the new cut on spatial frequency in pixels as 0.175/w
resolution (w
resolution is displayed below the spatial resolution) or
set the 2D DFT Butterworth BP filter the
unit of w parameter to
cycles/mm.
-
 Threshold:
Bottom at Pixel value = 0 (Threshold from local max/min=None):
this removes negative values created by the high pass filter.
Threshold:
Bottom at Pixel value = 0 (Threshold from local max/min=None):
this removes negative values created by the high pass filter.
-
 Wiener
Filter Smooth: adaptive filter for noise removal. Increase noise
level for stronger noise removal. Decrease noise level if
mitochondria are smudged.
Wiener
Filter Smooth: adaptive filter for noise removal. Increase noise
level for stronger noise removal. Decrease noise level if
mitochondria are smudged.
-
 Set
scaling/color and
Set
scaling/color and
 Write
back scaled values: values between 0-99.85. The Max
percentile 99.85 sets the sensitivity of the segmentation
operation below. Increase this value to have fewer segments.
Decrease this value to have more segments.
Write
back scaled values: values between 0-99.85. The Max
percentile 99.85 sets the sensitivity of the segmentation
operation below. Increase this value to have fewer segments.
Decrease this value to have more segments.
-
 Threshold:
Mask below Pixel value = 100 (Threshold from local
max/min=None): this suppresses background. Increase this
value if mitochondria look overflowing.
Threshold:
Mask below Pixel value = 100 (Threshold from local
max/min=None): this suppresses background. Increase this
value if mitochondria look overflowing.
-
 Set
Segmentation Classifiers: Limit Type=Max:
segments with parameters larger than the given values are
rejected (erased from the image). Zero means that the given
parameter is not looked. (no classifiers are set in the example)
Set
Segmentation Classifiers: Limit Type=Max:
segments with parameters larger than the given values are
rejected (erased from the image). Zero means that the given
parameter is not looked. (no classifiers are set in the example)
-
 Set
Segmentation Classifiers: Limit Type=Min:
segments with parameters smaller than the given values are
rejected (erased from the image). Zero means that the given
parameter is not looked. (no classifiers are set in the example)
Set
Segmentation Classifiers: Limit Type=Min:
segments with parameters smaller than the given values are
rejected (erased from the image). Zero means that the given
parameter is not looked. (no classifiers are set in the example)
-
 Advanced
Segmentation:
Otsu by Series, Above, at
1, Threshold from local max/min=Bound
locally, Determine boundaries at 10%),
Method=Watershed, Connectivity=Inf. This performs
local maximum search and segmentation with the combination of locally adaptive thresholding
and the watershed algorithm. In the segmented image segments are
marked with different values and frames are independent from
each other.
Advanced
Segmentation:
Otsu by Series, Above, at
1, Threshold from local max/min=Bound
locally, Determine boundaries at 10%),
Method=Watershed, Connectivity=Inf. This performs
local maximum search and segmentation with the combination of locally adaptive thresholding
and the watershed algorithm. In the segmented image segments are
marked with different values and frames are independent from
each other.
-
 Rename
Image to "Segments"
Rename
Image to "Segments"
- Showing and Exporting data (manually execute procedures, or
program it into Pipeline)
- Select Plotting/
 Plot
Intensities Corresponding to Segments: Type=Mean,
all other parameters are No.
Plot
Intensities Corresponding to Segments: Type=Mean,
all other parameters are No.
- Select the image "Segments" as
Image A. Select
the Optical Flow image (or any of the intensity images) as
Image B and press process
 in the tool bar. A plot window appears with circles
indicating values in Image B in overlapping pixels for each
segment of Image A. Segments are independent between frames
(i.e. are not tracked), therefore the Time Continuous
Segments parameter was set to No. The y-axis is labeled
with 'F' regardless the modality/unit of the measured quantity.
If not all images appear as Image B, use the
link
in the tool bar. A plot window appears with circles
indicating values in Image B in overlapping pixels for each
segment of Image A. Segments are independent between frames
(i.e. are not tracked), therefore the Time Continuous
Segments parameter was set to No. The y-axis is labeled
with 'F' regardless the modality/unit of the measured quantity.
If not all images appear as Image B, use the
link
 tool.
tool.
- Use the context menu (right
click) of the Plot window to save or copy data. If data is
copied to Excel, beware that Excel will handle only 253 columns,
so segments.
- Constraining segment evaluation to a ROI
- Draw a region ROI (on any of
the images because all are linked
by this time) with the
 toolbar button, encircling some of the segments.
toolbar button, encircling some of the segments.
- In the Plotting/
 Plot
Intensities Corresponding to Segments and
change the Constrain to Active ROI
parameter to Yes.
Plot
Intensities Corresponding to Segments and
change the Constrain to Active ROI
parameter to Yes.
- Perform plotting as above.
- Working with exported velocities
- Image Analyst MKII does not support statistical
evaluation of fluorescence or velocity data, therefore data
is exported as TAB delimited text file or by clipboard
copy/paste operation from the Plot Window.
- Use a statistical software capable of calculating
histograms and importing TAB delimited text files transposed
(e.g. Graphpad Prism or Sigmaplot)
- To determine number (or percentage) of fast moving or
stationary mitochondria calculate histograms. Mitochondrial
velocities typically show continuous distribution, therefore
the threshold between stationary and moving mitochondria has
to be arbitrarily determined.
 |
 |
 |
 |
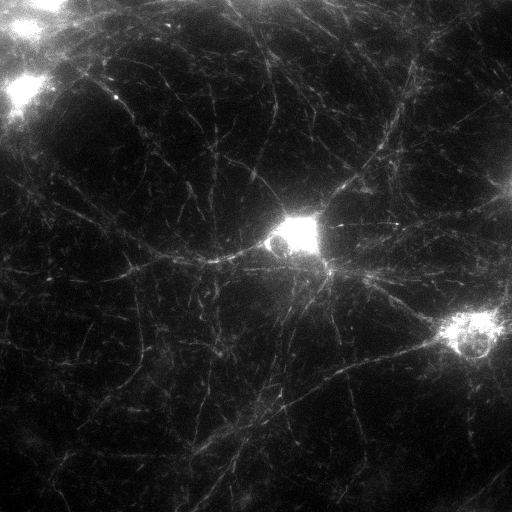
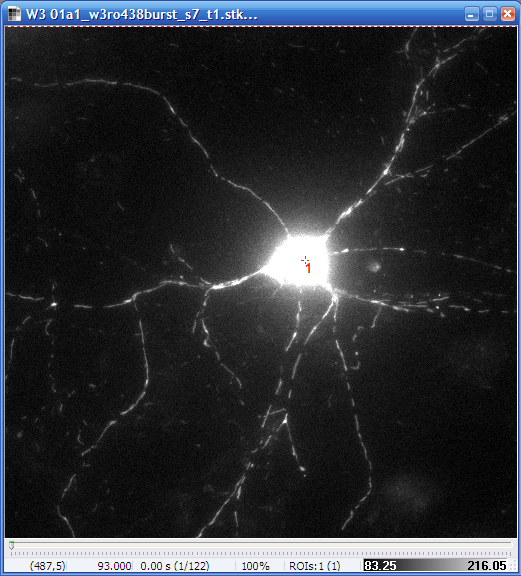
| Original, minimum intensity
projection image of the Optical Flow recording (GFP) |
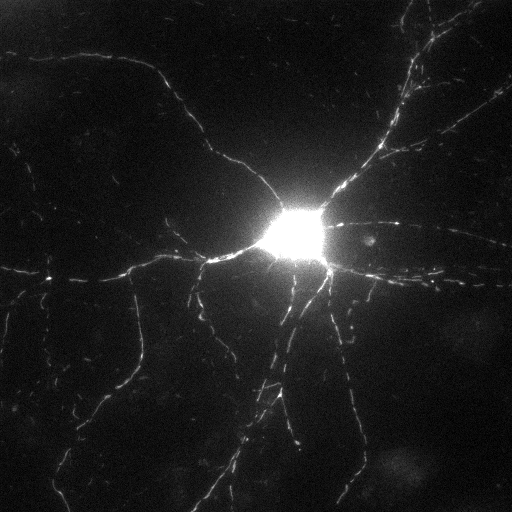
Filtered and smoothed image |
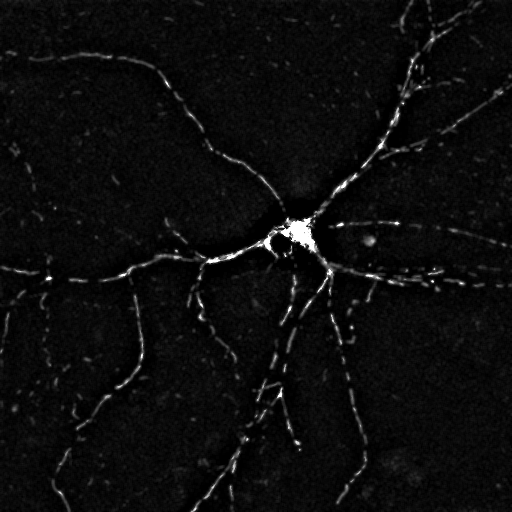
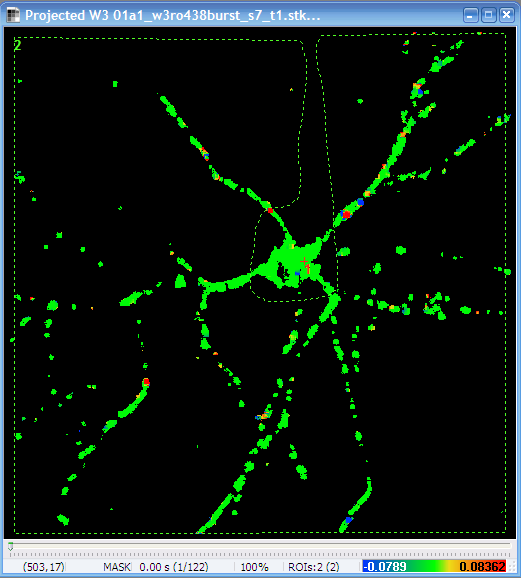
Segmented Image. Each segment is
marked by a different color.
Use the
 toolbar icon to explore basic parameters of the
segments by moving the mouse over the image.
toolbar icon to explore basic parameters of the
segments by moving the mouse over the image. |
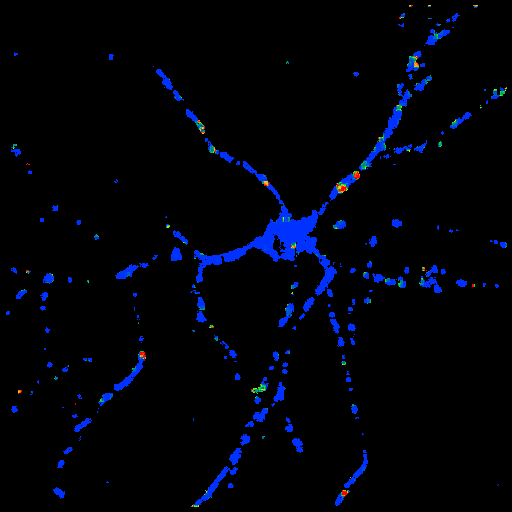
Optical Flow Image |
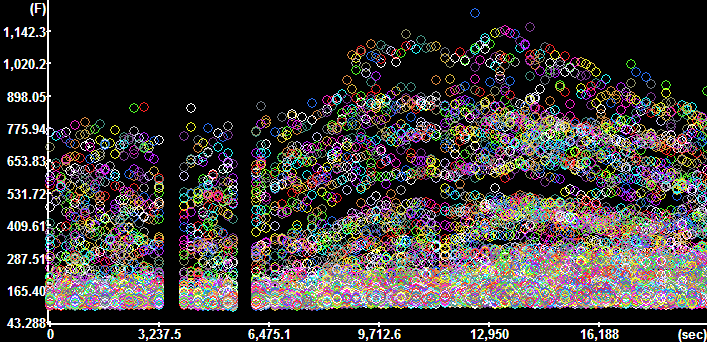 |
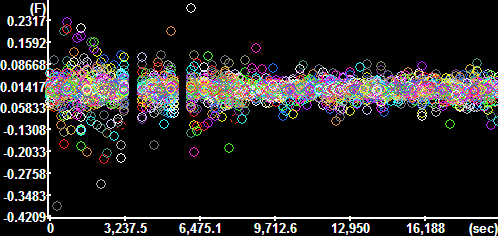
 Plot
Segment Intensities for the GFP channel (Time
Continuous segments=No, Constrain to active ROI=No) Plot
Segment Intensities for the GFP channel (Time
Continuous segments=No, Constrain to active ROI=No)
Each circle marks the value corresponding to and
individual segment.
The y-axis is scaled in fluorescence intensity. |
 |
 |
 Plot
Segment Intensities for the TMRM channel (Time
Continuous segments=No, Constrain to active ROI=No) Plot
Segment Intensities for the TMRM channel (Time
Continuous segments=No, Constrain to active ROI=No)
 |
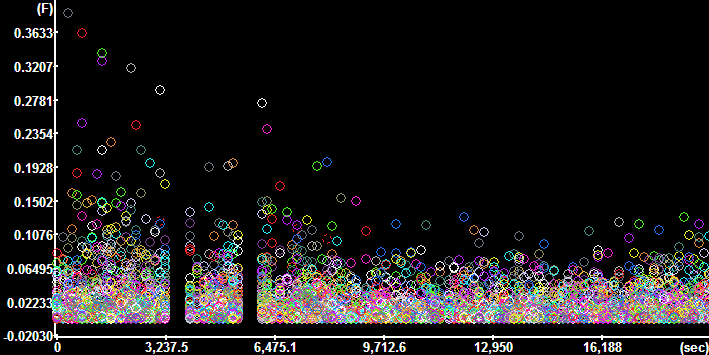 |
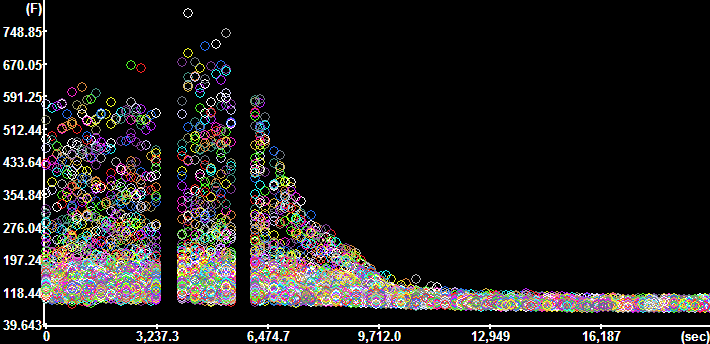
 Plot
Segment Intensities for the Optical Flow channel
(Time Continuous segments=No, Constrain to active
ROI=No). Plot
Segment Intensities for the Optical Flow channel
(Time Continuous segments=No, Constrain to active
ROI=No).
The y-axis is scaled in mm/sec. |
"mitochondrial
segmentation wide field.ips"
 |
Working with anterograde/retrograde motion using mean
radial velocities
- If using Multi-Dimensional Open dialog perform Optical flow loading as
follows:
- Use the
 point ROI tool to mark the
center of the cell.
point ROI tool to mark the
center of the cell.
- In the Multi-Dimensional Open dialog Processing set Optical Flow. In the
Optical Flow panel set the
Projection ROI parameter as number of the point
ROI.
- Set Output as Absolute value of Projected Vectors:
Yes
- Press Open.
- If using simple time lapse recordings, load the original
(raw) fluorescence time lapse image sequence.
- Use the
 point ROI tool to mark the
center of the cell.
point ROI tool to mark the
center of the cell.
- In the parameters of the
 Optical
Flow set Output as Absolute value of Projected Vectors=Yes
Optical
Flow set Output as Absolute value of Projected Vectors=Yes
- Perform Optical Flow calculation, and Block filtering of
the raw fluorescence images as detailed in the beginning of the
protocol.
 |
 |
| |
 |
|
Original image
with the point ROI in the soma |
Radial velocity
image (note the +/- scaling in the bottom)
The y-axis is scaled in mm/sec. |
Working with velocity vectors and transport/wiggling motion
- In the parameters of the
 Optical
Flow set Output as X and Y components of vectors=Yes
and Output as Absolute value of vectors=Yes
Optical
Flow set Output as X and Y components of vectors=Yes
and Output as Absolute value of vectors=Yes
- Using Plotting/
 Plot
Intensities Corresponding to Segments (Type=Mean,
all other parameters are No) and transfer
velocities corresponding to mean velocities, X and Y vector
components to statistics/spreadsheet software. Working with
or plotting vectors is not supported by Image Analyst MKII.
Plot
Intensities Corresponding to Segments (Type=Mean,
all other parameters are No) and transfer
velocities corresponding to mean velocities, X and Y vector
components to statistics/spreadsheet software. Working with
or plotting vectors is not supported by Image Analyst MKII.
- To obtain the wiggle ratio in the statistics/spreadsheet
software perform the following calculation:
- The wiggle ratio1 is the ratio of the
mean of absolute vectors over the absolute value of the
mean vector
- So calculate first the absolute value of mean vector
as SQRT(SQR(X-component)+SQR(Y-component))
- And divide the absolute optical flow with the above
result
- This results the wiggle ratio which has a value of
~1 for linear transport movement and is larger for
wiggling movement .
 |
 |
 |
 |
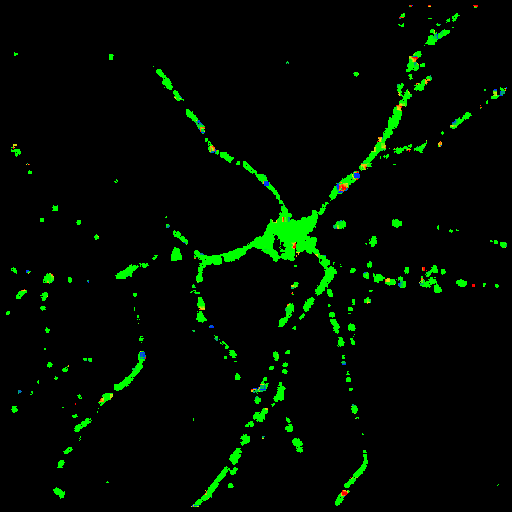
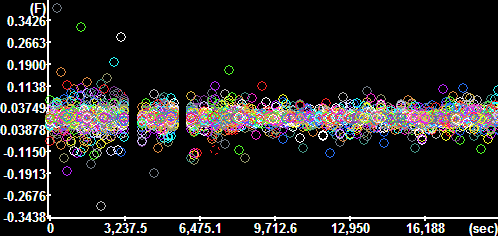
X component of the velocity vectors
The y-axis is scaled in mm/sec. |
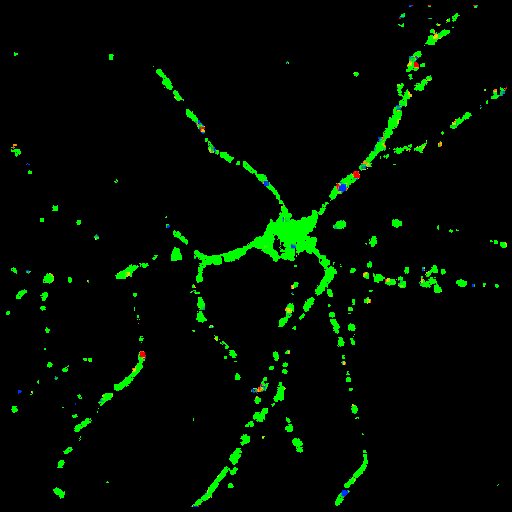
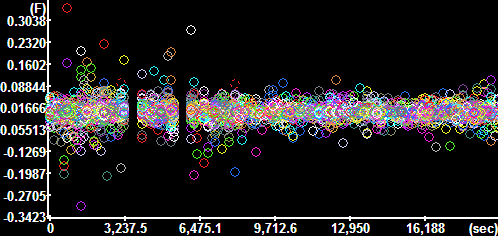
Y component of the
velocity vectors
The y-axis is scaled in mm/sec. |
Protocol by Akos A. Gerencser 03/07/2010 V1.0
References
1.
Gerencser
A. A. and Nicholls D. G. (2008) Measurement of Instantaneous
Velocity Vectors of Organelle Transport: Mitochondrial Transport and
Bioenergetics in Hippocampal Neurons. Biophys J. 2008 Sep
15;95(6):3079-99.







![]()