This protocol describes how to conduct a mitochondrial reactive oxygen species (ROS) measurement in adherent cells using fluorescence microscopy of MitoSOX (TPP-dihydroethidium) and image analysis in Image Analyst MKII. ROS levels are expressed as the rate of MitoSOX oxidation in %/sec normalized to MitoTracker Deep Red intensity (simple approach) or TMRM intensity (double normalization approach). The protocol is applicable to fluorescence microscopes capable of low-light level, time-lapse imaging. The normalization cancels the effects of differing cell size, density, geometry, and plasma and mitochondrial membrane potential-sensitive MitoSOX accumulation, that are otherwise a major confounding factors of the MitoSOX assay. Microscope settings for recording the superoxide-specific OH-ethidium fluorescence is given below. Disclaimer: we think that this approach is the available most precise one for fluorescence detection of MitoSOX oxidation, but even with using the superoxide-specific fluorescence excitation peak of OH-ethidium, and using the described normalization, the technique is not quantitative for superoxide, because of the complexity of the redox chemistry of dihydroethidium (see Zielonka and Kalyanaraman 2010, PMID: 20116425). Note that no blue fluorescence (460nm) emission from not-yet-oxidized MitoSOX can not be detected in contrast to DHE, therefore the normalization cannot be performed as simply as for DHE.
Image Analyst MKII supports this assay with background subtraction algorithms for low light level imaging, image registration, background masking, ROI graphing and rate calculations. These functionalities are integrated into a single, click-and-run pipeline specialized for this assay.
Equipment
Microscopy requirement:
The assay works with wide field (epifluorescence) and confocal microscopy. The epifluorescence microscope has to be capable of low-light level time lapse imaging, e.g. equipped with a fast shutter and a sensitive monochromatic camera. Confocal microscopes trivially work in low-light level mode. The image acquisition software must record accurate time stamps for optional rate calculation as 1/s. The assay protocol is given assuming that wide field microscopy and assay automation is used to capture multiple conditions in the same time.
Wide field microscopy:
Filter set: (notably, emission peaks are broad and other similar filters may work as well. This table also contains optional components)
| Filter | Spectrum (center/bandwidth nm) | Supplier & Cat. # | Note |
| MitoSOX OH-ethidium exciter (optional) | 390/40 | Semrock FF01-500/24-25 | individual stock filter, same as the DAPI exciter |
| TMRM (and optionally MitoSOX) exciter | 543/22 | standard Cy3 cube | |
| MitoSOX dichroic | 409 (or a bit higher, e.g. 495) | e.g. Semrock FF409-Di03-25x36 | standard Fura2 cube component |
| TMRM dichroic | 562 | Semrock FF562-Di03-25x36 | standard Cy3 cube |
| MitoSOX and TMRM emitter | 588/21 | Semrock FF02-588/21 -25 | individual stock filter, could be other similar |
| Standard Cy5 cube for MitoTracker Deep Red | 628/40 -> 692/40 | standard Cy5 cube |
The recommended setup: MitoSOX OH-ethidium is measured at 390nm -> 588nm. In the absence of a 390nm filter MitoSOX can be measured at 543nm->588nm in the same channel as TMRM (MitoSOX and TMRM are in different wells).
Confocal microscopy:
Two-photon microscopy:
| Reagent | Stock Concentration | Solvent | Storage | Supplier & Cat. # | Notes |
| Glucose | 1 M | H2O | RT | Sigma | sterile filter |
| Na-tetraphenylborate | 100 mM to 1 M | H2O | diluted stock is not stored | Sigma | multiple freeze thaw is OK |
| MitoSOX Red | 2 mM | DMSO | -20C, in anoxic bag | Molecular Probes, M36008 | freshly dissolve or use within a week |
| TMRM (optional) | 50 µM | DMSO | -20C | Molecular Probes, #M36008 | multiple freeze thaw is OK |
| MitoTracker Deep Red | 20-100 µM | DMSO | -20C | Molecular Probes, M22426 | multiple freeze thaw is OK |
| Zosuquidar (optional) | 25 mM | DMSO | -20C | MedKoo or Sigma | multiple freeze thaw is OK |
| IM (imaging medium) | see composition below | H2O | RT or 4C | sterile filter |
Imaging medium (IM):
| Substance | mM |
| NaCl | 120 |
| KCl | 3.5 |
| CaCl2 | 1.8 |
| MgCl2 | 1 |
| KH2PO4 | 0.4 |
| TES | 20 |
| NaHCO3 | 5 |
| Na2SO4 | 1.2 |
Note: Set pH to 7.4 using NaOH at 37ºC. Store medium aliquoted
in 50ml conicals at RT.
Assay protocol (given for LabTek 8-well chamber)
Handling and loading of cell cultures with MitoSOX
This assay needs two wells of cell culture for each condition in a multiwell chamber or plate. One will be used for the measurement of the MitoSOX rate and the other to measure the TMRM intensity as normalization factor. Culture cells on optical quality plastic bottom or glass bottom plates with equal density when several cell lines are compared. For robustness cells are recommended at 90% confluence on the plates, attached firmly and flattened down. Cell loss is expected due to washes therefore low cell density is not recommended. A confluent culture will prevent accurate background subtraction during data analysis.
In a conical warm up sufficient amount of IM (e.g. 20ml for an experiment in a LabTek 8-well chambered coverglass)
Supplement all of the IM with and sterile filter:
zosuquidar 1 µM (optional)
1 µM Na-tetraphenylborate
BSA 0.4% (can be used with this assay, optionally)
glucose 0 to 25mM depending on the experiment - by default match the composition of the culturing media
Note: Zosuquidar is a multi drug resistance protein (MDR) inhibitor and it may be used for comparison of specimens with different probe accumulation due to pumping. Note: that sterile filtering is to remove debris from the medium that may interfere with fluorescence imaging.
Add 5 nM MitoTracker Deep Red, to 4 ml of supplemented IM (immediately before steps 5 and 6)
Wash cell once with supplemented IM. Note: it is a good practice to keep cultures and media warm at 37ºC at all times.
Add the MitoTracker containing, supplemented IM to the wells (400ul/well for LabTek 8-well chamber) and incubate cultures for 45 minutes at 37ºC (non-CO2 incubator).
Meanwhile add 0.5 µM MitoSOX to 2 ml of supplemented IM and 10nM TMRM to another 2ml (immediately before steps 8 and 9)
Gently aspirate the probe-containing medium and wash once with supplemented IM.
Transfer the culture to the microscope.
Imaging
Channel setup:
MitoTracker Deep Red (CY5 cube)
TMRM 543nm->588nm
MitoSOX OH-ethidium 390->588nm
Optional channel setup:
MitoTracker Deep Red (CY5 cube)
TMRM and MitoSOX ethidium 543nm->588nm
Position setup:
Each condition must include a MitoSOX and a TMRM loaded well.
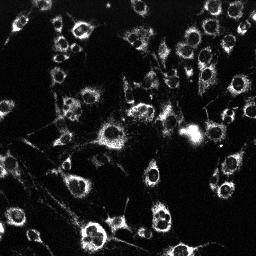
Set illumination intensity to see a very grainy, low light level image of the cells, barely distinguishable from the background (middle of top row in the figure below). Use high binning or low scan resolution if possible.
Record a time lapse for 20 min with 2 min frame intervals.
 |
 |
 |
 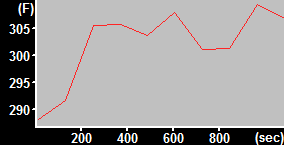 |
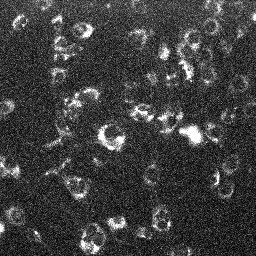
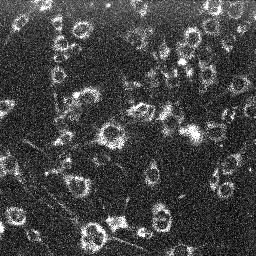
| MitoTracker Deep Red + MitoSOX stained C2C12 cells. From left to right: CY5 channel, 390->588nm, 543nm->588nm | MitoSOX rate (top) and MitoTracker Deep red intensity (bottom) | ||
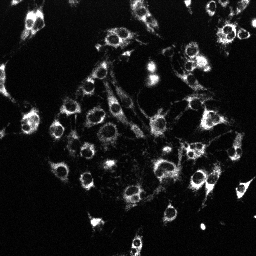 |
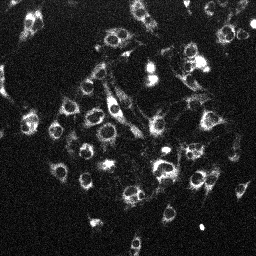 |
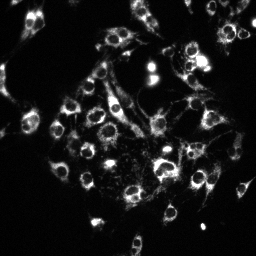 |
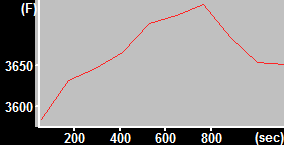  |
| MitoTracker Deep Red + TMRM stained C2C12 cells (sister well). From left to right: CY5 channel, 390->588nm, 543nm->588nm | TMRM intensity (top) and MitoTracker Deep red intensity (bottom) | ||
| Finally, the normalized rate is given by (MitoSOXtop rate / MTDRtop intensity) / (TMRMbottom intensity / MTDRbottom intensity). | |||
Image analysis in Image Analyst MKII
Please download the analysis pipeline an place it into the Documents\Image Analyst\My Pipelines\ folder. Press Edit/Rescan Pipelines Folder, if adding the new pipeline to the folder while Image Analyst is running.
Open the recording (![]() ).
).
Activate the downloaded "MitoSOX to MitoTracker Deep Red rates 3 channels" or "MitoSOX to MitoTracker Deep Red rates 2 channels" pipeline in the Pipelines main menu point. Note: this pipeline calculates the rate of increase in MitoSOX fluorescence intensity (output channel 1) and intensities of the TMRM channel (output channel 2) and the MitoTracker Deep Red channel (output channel 3). The image series is registered, background is subtracted, and cell-free areas, determined in a maximum intensity projection image are masked. The output is based on the mean fluorescence intensity of all cells in the view field.
Adjust the pipeline parameters:
Channel #: Make sure that channel numbers listed here match the actual recording
Background Level (percentile): Use a smaller background level (10-30 percentile) for confluent cultures and ~50 percentile for sparse cultures. Percentiles below 5-10 may increase noise.
Mask sensitivity (0.2-1.5): This sets the amount of background masking. At higher values only the brighter parts of the cells are measured, and more background is omitted from the mean fluorescence intensity.
Range for rate measurement: [important] Set the frame range where to measure rates and intensities, ideally from 1 to the last frame (e.g. 1-10).
Calculate rates for MitoSOX: Normally yes, turn it off to see whether the increase of MitoSOX fluorescence is linear. (you can also toggle this in the context menu of the Plot Window).
For the 3 channel (MitoSOX OH-ethidium setup):
Load the images and run
the pipeline by pressing
![]() .
Note: optionally choose to process the the whole multi-position
recording. Note: see a typical
result above.
.
Note: optionally choose to process the the whole multi-position
recording. Note: see a typical
result above.
In the Excel Data Window:
For each MitoSOX:MitoTracker Deep Red position calculate the ratio of Ch1_ROI1 and Ch3_ROI1 in an empty column
For each TMRM:MitoTracker Deep Red position calculate the ratio of Ch2_ROI1 and Ch3_ROI1 in an empty column
Finally, for each pair of sister cultures with MitoSOX and TMRM divide the two ratio values (form #1 and #2 above).
Save results in the Excel Data Window using the File/Save Excel Data Window main menu point.
For the 2 channel (MitoSOX OH-ethidium setup):
Calculate rates for MitoSOX or TMRM: set it yes
Run the pipeline for the MitoSOX wells using
the pull down menu of the
![]() button and selecting the appropriate range in the 'Run on
Partial Plate' menu point.
button and selecting the appropriate range in the 'Run on
Partial Plate' menu point.
Calculate rates for MitoSOX or TMRM: set it no
Load the images and run the pipeline for the TMRM wells using the pull down menu of
the
![]() button and selecting the appropriate range in the 'Run on
Partial Plate' menu point.
button and selecting the appropriate range in the 'Run on
Partial Plate' menu point.
In the Excel Data Window:
For each position calculate the ratio of Ch1_ROI1 and Ch2_ROI1 in an empty column
Finally, for each pair of sister cultures with MitoSOX and TMRM divide the above ratio values.
Save results in the Excel Data Window using the File/Save Excel Data Window main menu point.
Protocol by Irina by Irina Perevoshchikova and Akos A. Gerencser
11/12/2015
V1.0 ![]()