Measurement of mitochondria to cell volume fraction (V3.1)
This protocol describes how to measure mitochondria:cell volume fractions using a confocal microscope and image analysis in Image Analyst MKII. The assay was tuned to result similar volume fractions to electron microscopic stereological measurements when the below specified optical configuration is used for image acquisition. The technology is based on segmentation and binarization of high-resolution confocal micrographs of a cytosolic and a mitochondrial stain.
Sample preparation, reagents
Image acquisition with Zeiss LSM780 ZEN 2011, 2012 and with or without Zeiss LSM Multi Time Series PLUS version 24 or 35 module with Definite Focus
See links to previous versions of this protocol for older systems on the bottom of the page.
The aim is to record best possible quality, slightly oversampled images at evenly spaced z-coordinates from the bottom to the top of the culture. Due to strong photo-toxicity and photo-bleaching it is not possible to use z-stacking, but different x,y-coordinates of the culture are scanned systematically at incrementing the z-focus. To automate this, the Zeiss LSM Multi Time Series dialogue is used with overriding Z-offset values with an Microsoft Excel / Windows regedit trick. See also recording of volume fraction data without using the Multi Time Lapse module. Microscope Settings are given below for Zeiss LSM 780, ZEN2011 software with Multi Time Series PLUS, ZEN 2011-24 module equipped with Definite Focus. Here the protocol is given for an LSM780 equipped with a spectral detector and 561nm diode laser. See 'Manual image acquisition' below if using other microscope or not having the Multi Time Lapse module.
|
|
|
|
|
Saving tab |
Acquisition tab |
Grid tab (described below) |
|
Autofocus |
Timing Tab (missing) |
Windows Registry Editor Version 5.00
[HKEY_CURRENT_USER\Software\Carl Zeiss Jena GmbH\UI\AutoTime\VolumeRatio]
[HKEY_CURRENT_USER\Software\Carl Zeiss Jena GmbH\UI\AutoTime\VolumeRatio\Block1]
1. Focus an area in the sample that is not going to be recorded.
2. Set up laser powers and gains to result no saturation of images. Saturated pixels prevent accurate analysis.
3. Save the adjusted ‘calcein-MTR’ configuration.
4. In the ZEN press Locate and switch to eyepiece / transmitted light
5. If doing post-hoc immunocytochemistry, set the top edge of the well into the center of the view field, go to the Acquisition tab and zero the focus by pressing 'Manually' in the Focus panel, and zero the stage by pressing 'Set zero' in the Stage panel
6. Go back to the Multi Time Series
7. Search for a 50-100 cells under eyepiece, center each in the view field and press 'Add NEXT'.
8. Optionally cycle around all the selected cells to see that they are centered, and there are no cells imaged more than once.
9. Move to the first position
10. In the ZEN turn off 488 and set 561 laser to 5% power and using live scan focus the lowest plane where mitochondria are sharply visible. Stop scanning.
11. In the Multi Time Series, Autofocus tab, press ‘Find Z Offset’. [The offset will show zero, but the focusing is now initialized.]
12. Save the above settings in the Saving tab Store/Apply Recipe under the ‘VolumeRatio’ name. Wait until done.
13. In the Windows Explorer double click volumeratio.reg and OK to enter its contents to the system Registry.
14. Use the Store/Apply button to Apply parameters into registry under the name of “VolumeRatio”.
15. In the Acquisition tab set Scan Configuration to 'calcein-MTR': and press Apply to All Locations.
16. Verify other settings as above given in the Multi Time Series, including file name and folder.
17. Press Start.
18. If doing post-hoc immunocytochemistry, after finished imaging in the Saving tab Store the recipe under a unique name. This will store the image coordinates for later revisiting.
1. Focus an area in the sample that is not going to be recorded.
2. Set up laser powers and detector gains to result no saturation of images. Saturated pixels prevent accurate analysis.
3. Save the adjusted ‘calcein-MTR’ configuration.
4. In the ZEN press Locate and switch to eyepiece / transmitted light
5. If doing post-hoc immunocytochemistry, set the top edge of the well into the center of the view field, go to the Acquisition tab and zero the focus by pressing 'Manually' in the Focus panel, and zero the stage by pressing 'Set zero' in the Stage panel
6. Go back to the Multi Time Series
7. Select a homogeneous area of in each well to be imaged and press 'Add NEXT'. Note: you may press the clear all locations button before this step.
8. Optionally cycle around all the selected wells to see that they are still in focus.
9. Optionally save the settings.
10. In the Grid tab set 10 for X and 5 for Y and 100 microns for both Grid steps. [Note that these numbers have to be entered even if they are already there by default, because the dialogue tends to take zero grid steps otherwise (this was seen at version 24).]
11. Check Meander mode and press Create Multi Grids.
12. Verify that you have now 50 times the number of wells selected locations.
13. Move to the first position (just press Next Loc)
14. In the ZEN turn off 488 and set 561 laser to 5% power and using live scan focus the lowest plane where mitochondria are sharply visible. Stop scanning.
15. In the Multi Time Series, Autofocus tab, press ‘Find Z Offset’. [The offset will show zero, but the focusing is now initialized.]
16. Save the above settings in the Saving tab Store/Apply Recipe under the ‘VolumeRatio’ name. Wait until done. [Note: It is important to used this configuration name for the procedure below.]
17. Adjust the number of rows in the ‘volumeratioDefiniteFocus.xls’ to match (or exceed) the number of total positions, and copy columns to the ‘volumeratio.reg’ as above described.
18. In the Windows Explorer double click ‘volumeratio.reg’ and OK to enter its contents to the system Registry.
19. Use the Store/Apply button to Apply parameters into registry under the name of “VolumeRatio”.
20. In the Acquisition tab set Scan Configuration to 'calcein-MTR': and press Apply to All Locations.
21. Verify other settings as above given in the Multi Time Series, including file name and folder.
22. Press Start.
23. Keep an eye on the microscope, checking whether the Definite Focus correctly engages as the acquisition moves from well to well. If fails reload the configuration saved in #9 and repeat from #10 using the remaining well positions.
24. If doing post-hoc immunocytochemistry, after finished imaging in the Saving tab Store the recipe under a unique name. This will store the image coordinates for later revisiting.
5. Recording post-hoc immunocytochemistry (requiring the Multi Time Series PLUS module of the Zeiss ZEN software)
1. In the ZEN load the previously defined ‘counterstain’ configuration. If not yet defined do the following:
1. load ‘calcein-MTR’ configuration, and change the Light Path and Channels to match the fluorescence of the label. Usually calcein and Mitotracker Red are washed out by this time or outshined by the staining.
2. Keep the same Acquisition mode settings, however Faster speed and Bi-directional scanning may be used.
3. Save the new configuration as ‘counterstain’ (arbitrary name).
2. Press Locate and switch to eyepiece / transmitted light and set the top edge of the well into the center of the viewfield, go to the Acquisition tab and zero the focus by pressing 'Manually' in the Focus panel, and zero the stage by pressing 'Set zero' in the Stage panel
3. Go back to the Multi Time Series and under Saving tab Store/Apply Recipe load the previously stored coordinates. [Note, that stored coordinates need to be loaded after pressing ‘Set zero’ above.]
4. In the ZEN load the live cell calcein-MTR recording. You may split the screen to see live image and the original recording in the same time.
5. In the Multi Time Series select any position and using Live scan in the ZEN, try to find the same cell compared to the live recording, by dragging the scan area in the Acquisition mode panel. [Note, that tiled acquisition is also an option to find the same cell]
6. If not finding the cell, the zero position may need to be adjusted, and coordinates re-loaded.
7. If the Live scan image matches the previously recorded calcein-MTR image, stop scanning and save the ‘counterstain’ configuration.
8. Move to the first position
9. Using live scan focus the lowest plane where the cell is sharply visible. Stop scanning.
10. In the Multi Time Series, Autofocus tab, press ‘Find Z Offset’. [The offset will show zero, but the focusing is now initialized.]
11. Save the above settings in the Saving tab Store/Apply Recipe under the ‘VolumeRatio’ name. Wait until done. [Note: It is important to use this configuration name for the procedure below.]
12. In the Windows Explorer double click the previously used ‘volumeratio.reg’ and OK to enter its contents to the System Registry.
13. Use the Store/Apply button to Apply parameters into registry under the name of “VolumeRatio”.
14. In the Acquisition tab set Scan Configuration to ‘counterstain’ and press Apply to All Locations.
15. Verify other settings as above given in the Multi Time Series, including file name and folder.
16. Press Start. [Note, if split-screen was used, you can side-be-side scroll the live-cell recording to verify the match.]
Analysis in Image Analyst MKII (Version 3.0.0 and above)
Use Image Analyst MKII to determine areas corresponding to mitochondria and the whole cell by using adaptive thresholding.
Four pipelines are available under the Pipelines/Morphological Measurements/Applications main menu point, with slightly different functionalities:
Adjustments necessary to run the pipeline
Select the pipeline in the main menu, and look the parameter bar (below). Adjustment may need to be done if using a different microscope configuration for recording than the above specified. The "Sensitivity scaling for the cytosolic stain (percentile)" parameter may need to be adjusted if cell density is very different or the cells have bright vacuoles.
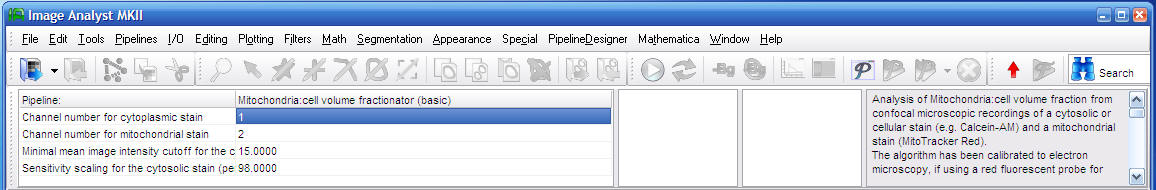
Parameters of the basic volume fractionator:
Channel number for ...: set the corresponding channel number as shown in the Multi Dimensional Open dialog.
Minimal mean image intensity cutoff for the cytoplasmic stain: The pipeline discards blank images to prevent biasing intensity rescaling of the image series. Such blank frames may occur in high z-planes, or if an automatically spread grid misses any cells. The mean intensity of an image containing minimal useful information needs to be set. To find this value create a whole-image ROI and plot mean intensities. Look typical values for empty frames, and set a slightly larger value.
Sensitivity scaling for the cytosolic stain (percentile): A lower value (slightly below 100) increases sensitivity. And estimates bigger cell size. Cells aggregating the probe in lysosomes need a lower percentile value to select cells and not lysosomes.
Additional parameters for the original volume fractionator:
Detector bit-depth for detection of saturation : provide the detector bit depth set before acquisition e,g, 8,12 or 16.
Additional parameters for the multiplexed volume fractionator:
Cell tracker threshold (%) : this threshold will be used to distinguish the two populations (percent of maximal fluorescence)
Spectral Unmix Coefficient Matrix for channel 2 and 3 : Spectral unmix matrix between Mitotracker Red and presumably deep red fluorescence multiplexer stain
Additional parameters for the basic with post-hoc staining volume fractionator:
Set channel numbers and lookup tables for the post-hoc staining. These images will appear next to the processed images helping manual masking of desired cell types.
Performing the analysis
Open the recording (up to 100 frames for basic processing and up to 50 frames for multiplexed or with post-hoc stain processing with 4GB or more RAM). If data was recorded in separate files use multiple file selection to open.
Run the pipeline
![]() .
Alternatively use the
.
Alternatively use the
![]() button to load and process.
button to load and process.
The pipeline will automatically load a template spreadsheet, fill in the data and calculate the volume fraction.
Save the contents of the Excel Data Window using the File/Save Excel Data main menu.
Analysis protocol with post-hoc immunostaining
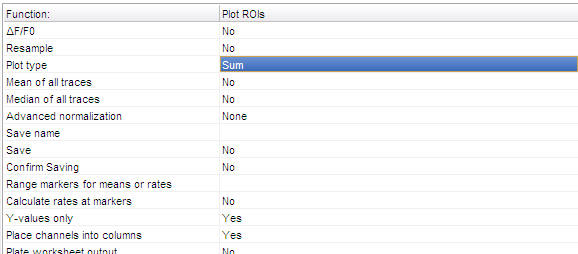
Note: the parameters not visible above are set to "No".
Protocol by Akos A. Gerencser 07/29/2015
V3.1 ![]()
Who to cite? This technique has been published here:
We used this technology in the following papers:
Using post-hoc immunostaining: